Greenplum 에 바이오파이썬 모듈을 설치하여 pl/python function 에서 바이오파이썬을 사용해 보도록 하자.
바이오파이썬 공식홈페이지 - http://biopython.org
Biopython · Biopython
Biopython See also our News feed and Twitter. Introduction Biopython is a set of freely available tools for biological computation written in Python by an international team of developers. It is a distributed collaborative effort to develop Python librarie
biopython.org

Procedural Languages 이란
2020/12/23 - [Database/Greenplum] - Greenplum - Procedural Languages 란?
Greenplum - Procedural Languages 란?
Greenplum Function 을 SQL만 이용하여 개발하는 것이아니고 Java, Python 등 언어를 선택하여 개발할 수 있게끔 해주는 기능이 Procedural Languages 이다. SQL의 한계를 넘어서 자바의 특정 패키지/라이브러리,.
wylee-developer.tistory.com
파이썬은 많은 모듈이 개발되어 있고 자유롭게 사용할 수 있으며 이 포스팅에서는 바이오파이썬 모듈을 설치해 보자.
바이오파이썬이란 분자생물학 계산에 자유롭게 사용하기 위한 파이썬 라이브러리로써 간단한 정보(?)는 위키백과를 참고
하지만 바이오파이썬 모듈을 사용하려면 해당 모듈을 Greenplum을 구성하고 있는 모든 노드에 설치해주어야 한다. (Greenplum 클러스터 구성이 6 Node 이면 6 Node에 전부 설치)
## 설치환경
- RedHat 7.5
- Greenplum 6.11.1
- Python 2.7.12
1. 설치된 파이썬 버전을 확인한다.(gpadmin 계정)
python --version
[gpadmin@mdw ~]$ python --version
Python 2.7.12
2. biopython-1.76 파일다운로드
biopython-1.6.x 은 pyhton 2.6 까지 지원사항이여서 설치가 불가능하고 python 2.7 이면 1.7.x 설치를 권고한다.
pypi.org/project/biopython/1.76/
biopython
Freely available tools for computational molecular biology.
pypi.org
biopython-1.76 파일다운로드
pypi.org/project/biopython/1.76/#files
biopython
Freely available tools for computational molecular biology.
pypi.org
접속 후 biopython-1.76.tar.gz 을 선택하여 설치파일 다운로드
3. 설치할 노드에 파일을 업로드 후 파일을 압축해제한다.
tar zxf biopython-1.76.tar.gz
[gpadmin@mdw opt]$ tar zxf biopython-1.76.tar.gz
[gpadmin@mdw opt]$ ll
total 15912
drwxr-xr-x 8 gpadmin gpadmin 4096 Dec 20 2019 biopython-1.76
-rw-r--r-- 1 gpadmin gpadmin 16283634 Dec 22 15:41 biopython-1.76.tar.gz
[gpadmin@mdw opt]$
4. 폴더 이동 후 파이썬 설치모듈 명령어 python stup.py build 명령어를 실행한다.
python setup.py build && python setup.py install
[gpadmin@mdw opt]$ cd biopython-1.76
[gpadmin@mdw biopython-1.76]$ python setup.py build && python setup.py install
==================================================================
WARNING: Biopython will drop support for Python 2.7 in early 2020.
==================================================================
running build
running build_py
creating build
creating build/lib.linux-x86_64-2.7
creating build/lib.linux-x86_64-2.7/Bio
copying Bio/kNN.py -> build/lib.linux-x86_64-2.7/Bio
copying Bio/LogisticRegression.py -> build/lib.linux-x86_64-2.7/Bio
copying Bio/_utils.py -> build/lib.linux-x86_64-2.7/Bio
copying Bio/triefind.py -> build/lib.linux-x86_64-2.7/Bio
copying Bio/bgzf.py -> build/lib.linux-x86_64-2.7/Bio
copying Bio/NaiveBayes.py -> build/lib.linux-x86_64-2.7/Bio
copying Bio/File.py -> build/lib.linux-x86_64-2.7/Bio
copying Bio/MarkovModel.py -> build/lib.linux-x86_64-2.7/Bio
.
.
.
running build_ext
building 'Bio.Align._aligners' extension
creating build/temp.linux-x86_64-2.7
creating build/temp.linux-x86_64-2.7/Bio
creating build/temp.linux-x86_64-2.7/Bio/Align
gcc -pthread -fno-strict-aliasing -g -O2 -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC -I/usr/local/greenplum-db-6.11.1/ext/python/include/python2.7 -c Bio/Align/_aligners.c -o build/temp.linux-x86_64-2.7/Bio/Align/_aligners.o
gcc -pthread -shared build/temp.linux-x86_64-2.7/Bio/Align/_aligners.o -L/opt/python-2.7.12/lib -lpython2.7 -o build/lib.linux-x86_64-2.7/Bio/Align/_aligners.so
/usr/bin/ld: cannot find -lpython2.7
collect2: error: ld returned 1 exit status
error: command 'gcc' failed with exit status 1
[gpadmin@mdw biopython-1.76]$
gcc 관련된 에러가 발생한다.
파이썬은 주개발언어가 아니라 에러메세지를 잘못보던 와중 파이썬 모듈 설치시 test --offline 옵션으로 설치관련 상세로그를 확인할 수 방법이 있는 걸 찾아냈다. (와우!! 역시 구글신!!)
python setup.py test --offline
[gpadmin@mdw biopython-1.76]$ python setup.py test --offline
==================================================================
WARNING: Biopython will drop support for Python 2.7 in early 2020.
==================================================================
running test
Skipping any tests requiring internet access
Python version: 2.7.12 (default, Jul 22 2020, 00:35:02)
[GCC 4.8.5 20150623 (Red Hat 4.8.5-39)]
Operating system: posix linux2
test_Ace ... ok
test_Affy ... skipping. Install NumPy if you want to use Bio.Affy.CelFile
test_AlignIO ... ERROR
test_AlignIO_ClustalIO ... ERROR
test_AlignIO_EmbossIO ... ERROR
test_AlignIO_FastaIO ... ERROR
.
.
.
<힌트메세지>
test Affy ... skipping. Install NumPy if you want to use Bio.Affy.CelFile
NumPy 모듈 설치가 필요하다고 나온다.!!!
biopython 설명페이지를 찾아보니 잘 설명되어 있다.(역시 매뉴얼을 잘 봐야 한다.;;;;;)
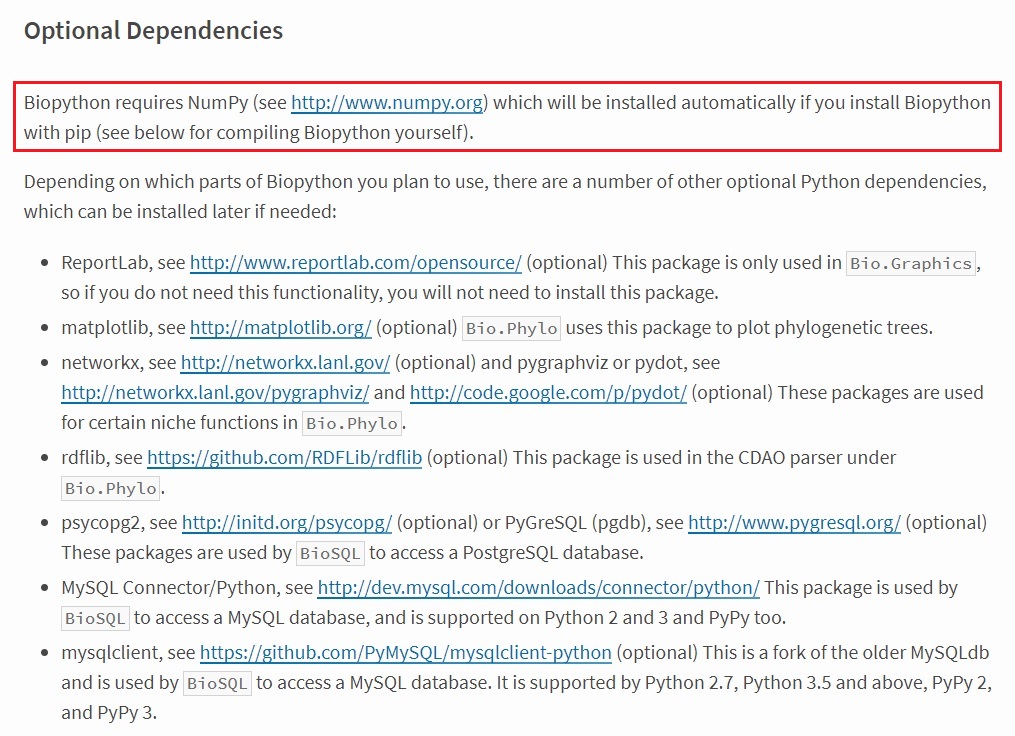
5. NumPy 1.16.0 - 파이썬 모듈 다운로드 및 설치
NumPy 모듈의 파이썬 2.7을 지원하는 마지막 버전은 v1.16.0 입니다. (v1.7.x 부터는 파이썬 3.x 만 지원한다.)
Release v1.16.0 · numpy/numpy
========================== NumPy 1.16.0 Release Notes This NumPy release is the last one to support Python 2.7 and will be maintained as a long term release with bug fixes until 2020. Support for ...
github.com
압축해제 후 설치
tar zxf numpy-1.16.0.tar.gz
cd numpy-1.16.0
python setup.py build && python setup.py install
[gpadmin@mdw opt]$ tar zxf numpy-1.16.0.tar.gz
[gpadmin@mdw opt]$ cd numpy-1.16.0
[gpadmin@mdw numpy-1.16.0]$ python setup.py build && python setup.py install
.
.
compile options: '-Inumpy/core/src/common -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/usr/local/greenplum-db-6.11.1/ext/python/include/python2.7 -c'
gcc: _configtest.c
gcc -pthread _configtest.o -o _configtest
success!
removing: _configtest.c _configtest.o _configtest.o.d _configtest
C compiler: gcc -pthread -fno-strict-aliasing -g -O2 -DNDEBUG -g -fwrapv -O3 -Wall -Wstrict-prototypes -fPIC
compile options: '-Inumpy/core/src/common -Inumpy/core/src -Inumpy/core -Inumpy/core/src/npymath -Inumpy/core/src/multiarray -Inumpy/core/src/umath -Inumpy/core/src/npysort -I/usr/local/greenplum-db-6.11.1/ext/python/include/python2.7 -c'
gcc: _configtest.c
_configtest.c:1:5: warning: conflicting types for built-in function ‘exp’ [enabled by default]
int exp (void);
^
.
.
.
정상적으로 설치가 되지 않고 위의 에러가 발생하면 python-devel.x86_64 rpm 을 설치해주어야 한다. (root 권한으로 rpm 설치)
yum list | grep python-devel
yum install -y python-devel.x86_64
[root@mdw opt]# yum list | grep python-devel
nautilus-python-devel.x86_64 1.2.2-2.el7 epel
nemo-python-devel.x86_64 3.6.0-3.el7 epel
python-devel.x86_64 2.7.5-68.el7 RHEL7.5
qscintilla-python-devel.noarch 2.8-1.el7 epel
[root@mdw opt]# yum install -y python-devel.x86_64
rpm 설치가 완료되었으면 다시 NumPy 설치
python setup.py build && python setup.py install
[gpadmin@mdw numpy-1.16.0]$ python setup.py build && python setup.py install
.
.
.
Installed /usr/local/greenplum-db-6.11.1/ext/python/lib/python2.7/site-packages/numpy-1.16.0-py2.7-linux-x86_64.egg
Processing dependencies for numpy==1.16.0
Finished processing dependencies for numpy==1.16.0
[gpadmin@mdw numpy-1.16.0]$
정상적으로 설치완료!!! - Finished
5. 다시 바이오파이썬 설치
[gpadmin@mdw numpy-1.16.0]$ cd /opt/biopython-1.76
[gpadmin@mdw biopython-1.76]$ python setup.py build && python setup.py install
.
.
.
running install_egg_info
Copying biopython.egg-info to /usr/local/greenplum-db-6.11.1/ext/python/lib/python2.7/site-packages/biopython-1.76-py2.7.egg-info
running install_scripts
==================================================================
WARNING: Biopython will drop support for Python 2.7 in early 2020.
==================================================================
[gpadmin@mdw biopython-1.76]$
6. 설치된 모듈 확인
python -c "help('modules')" | grep -i bio
[gpadmin@mdw ~]$ python -c "help('modules')" | grep -i bio
/usr/local/greenplum-db-6.11.1/ext/python/lib/python2.7/site-packages/Bio/Align/substitution_matrices/__init__.py:21: BiopythonExperimentalWarning: Bio.Align.substitution_matrices is an experimental module which may still undergo significant changes. In particular, the location of this module may change, and the Array class defined in this module may be moved to other existing or new modules in Biopython.
BiopythonExperimentalWarning)
/usr/local/greenplum-db-6.11.1/ext/python/lib/python2.7/site-packages/Bio/Crystal/__init__.py:44: BiopythonDeprecationWarning: Bio.Crystal has been deprecated, and we intend to remove it in a future release of Biopython. Please use Bio.PDB instead to parse NDB files.
" to parse NDB files.", BiopythonDeprecationWarning)
/usr/local/greenplum-db-6.11.1/ext/python/lib/python2.7/site-packages/Bio/KDTree/__init__.py:27: BiopythonDeprecationWarning: Bio.KDTree has been deprecated, and we intend to remove it in a future release of Biopython. Please use Bio.PDB.kdtrees instead, which is functionally very similar.
BiopythonDeprecationWarning,
/usr/local/greenplum-db-6.11.1/ext/python/lib/python2.7/site-packages/Bio/Statistics/__init__.py:15: BiopythonDeprecationWarning: Bio.Statistics has been deprecated, and we intend to remove it in a future release of Biopython.
"in a future release of Biopython.", BiopythonDeprecationWarning)
/usr/local/greenplum-db-6.11.1/ext/python/lib/python2.7/site-packages/Bio/codonalign/__init__.py:27: BiopythonExperimentalWarning: Bio.codonalign is an experimental module which may undergo significant changes prior to its future official release.
BiopythonExperimentalWarning)
/usr/local/greenplum-db-6.11.1/ext/python/lib/python2.7/site-packages/Bio/phenotype/__init__.py:100: BiopythonExperimentalWarning: Bio.phenotype is an experimental submodule which may undergo significant changes prior to its future official release.
BiopythonExperimentalWarning,
Bio asynchat importlib shelve
BioSQL asyncore imputil shlex
[gpadmin@mdw ~]$
에러메세지가 표시되지만 무시하자. 향후 버전에서 지원종료예정 경고메세지가 대부분이다.
7. 실행테스트
바이오파이썬 함수를 사용해서 정상적으로 설치가 되었는지 확인하자.
생물정보학자의 블로그 참고 - https://korbillgates.tistory.com/72
[바이오파이썬] 02. 바이오파이썬으로 할 수 있는 일들
안녕하세요 한주현 입니다. 오늘은 바이오파이썬으로 할 수 있는 일들에 대해서 말씀드리겠습니다. 시작하며 바이오파이썬은 생물학자들이 컴퓨터로 작업을 할 때 도움이 될 만한 파이썬 라이
korbillgates.tistory.com
vi bio-test.py
python bio-test.py
[gpadmin@mdw opt]$ vi bio-test.py
from Bio.Seq import Seq
my_seq = Seq("AGTAGAAAAABBBBTTT")
print(my_seq)
print(my_seq.alphabet)
print(my_seq.complement())
print(dir(my_seq))
:wq
[gpadmin@mdw opt]$
[gpadmin@mdw opt]$ python bio-test.py
AGTAGAAAAABBBBTTT
Alphabet()
TCATCTTTTTVVVVAAA
['__add__', '__class__', '__contains__', '__delattr__', '__dict__', '__doc__', '__eq__', '__format__', '__ge__', '__getattribute__', '__getitem__', '__gt__', '__hash__', '__imul__', '__init__', '__le__', '__len__', '__lt__', '__module__', '__mul__', '__ne__', '__new__', '__radd__', '__reduce__', '__reduce_ex__', '__repr__', '__rmul__', '__setattr__', '__sizeof__', '__str__', '__subclasshook__', '__weakref__', '_data', '_get_seq_str_and_check_alphabet', 'alphabet', 'back_transcribe', 'complement', 'count', 'count_overlap', 'encode', 'endswith', 'find', 'index', 'join', 'lower', 'lstrip', 'reverse_complement', 'rfind', 'rindex', 'rsplit', 'rstrip', 'split', 'startswith', 'strip', 'tomutable', 'transcribe','translate', 'ungap', 'upper']
[gpadmin@mdw opt]$
문제없이 실행되면 바이오파이썬을 정상적으로 설치가 완료된 것이다.
<끄읕>
'Database > Greenplum' 카테고리의 다른 글
| Greenplum 7 RoadMap - 예상기능정리 (0) | 2021.03.05 |
|---|---|
| Greenplum 6.14에서 ORCA(query optimizer) 성능향상 (0) | 2021.02.17 |
| Greenplum - Procedural Languages 란? (0) | 2020.12.23 |
| Greenplum PXF 란? (0) | 2020.12.16 |
| Greenplum minor 업그레이드하기(6.3.0->6.11.1) (0) | 2020.12.16 |


